Microorganisms are ubiquitous in geological environments, from the surface of the earth to the deepest layer of the ocean’s crust. Some microorganisms are useful, while others can be very harmful. In oil and gas production, microbiological activity is one of the biggest contributors to metal infrastructure corrosion. While there are some available solutions to mitigate microbiologically influenced corrosion (MIC), a big challenge is to make the connection between corrosion damage, such as pitting, and the activity of microorganisms.1
 While still used, traditional culture-based methods such as serial dilution or most probable number testing using “bug bottles” have failed to detect predominant troublesome microorganisms.2 These observed limitations have led to the adoption of molecular microbiological methods (MMM) to identify bacteria in oil field systems.3-4 MMM systematically links the type (species, family, or class) of microorganisms to corrosion and identifies the total microbiological population (dead, alive, and dormant) at the corrosion site.2 Since adopting MMM in oil and gas systems over 10 years ago, there is now a deeper understanding of how molecular tools can be used to identify MIC.5
While still used, traditional culture-based methods such as serial dilution or most probable number testing using “bug bottles” have failed to detect predominant troublesome microorganisms.2 These observed limitations have led to the adoption of molecular microbiological methods (MMM) to identify bacteria in oil field systems.3-4 MMM systematically links the type (species, family, or class) of microorganisms to corrosion and identifies the total microbiological population (dead, alive, and dormant) at the corrosion site.2 Since adopting MMM in oil and gas systems over 10 years ago, there is now a deeper understanding of how molecular tools can be used to identify MIC.5
The threat of MIC occurs across metallic oil and natural gas pipeline networks that are exposed to water.4 In MIC assessment specifically, several MMM (e.g., the quantitative polymerase chain reaction method using 16S rRNA universal primers and the Next Generation Sequencing method) are useful tools to quantify the abundance and diversity of many types of bacteria and archaea.6-7 Results enable the end user to identify thermophilic archaea, methanogens, sulfate-reducing bacteria (SRB), nitrogen fixers, and obligate anaerobes, to name a few examples.5 Some case studies, however, have highlighted challenges in interpreting data from MMM. Factors such as the time between sampling and analysis, operating conditions, and sample integrity can lead to erroneous results. Also, microbiological data interpretation in the context of the operating environment (e.g., temperature, salinity, or flow rate) is needed to form valid scientific conclusions.2,5
During the decade since these molecular tools were adopted, two major limitations have been identified. The first limitation is lack of established connectivity between MMM and corrosion data because there are no published methods to collectively assess other relevant information about MIC (e.g., operating conditions, system chemistry, and metallurgy). The second limitation is maintaining sample integrity prior to analysis. Operators in the field collect samples without knowledge of sterile techniques, and shipping from offshore platforms can take weeks to months.
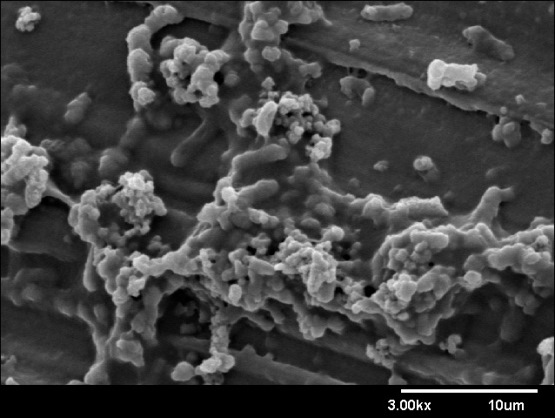
To overcome these limitations, advances in the application of MMM could occur through a systematic and organized movement that is inclusive of all participants (operators, vendors, and scientists).8 Solutions can be categorized into education, data management, and collaboration.
Educating operators in proper sterile techniques and sample collections is needed to reduce the likelihood of contamination and sample degradation. Training that focuses on sterile techniques to be used during sample collection could be provided in a classroom. Another option would be the development of a video tutorial that can be viewed on a mobile device for easy access when needed in the field.
Data management is also critical, particularly because of the large volume of data produced in metagenomics. Data could be stored on an online portal to record important categories such as field locations, operating conditions, and chemical or metallurgical variables. A numerical “MIC severity scale,” along with the identification of microorganisms to establish connections between molecular results and corrosion damage (e.g., biotic vs. abiotic corrosion), also would be helpful.
Additionally, collaboration is needed between researchers, academic partners, and start-up companies so information about new methodologies or technologies being developed can be shared. In the digital health industry, for example, portable medical devices are being developed for point-of-care technologies that can generate results of a blood test right at a patient’s bedside. Start-up companies in energy technology could develop sensors for a portable MIC monitoring kit or portable methodological panels.9 Having a device that can reliably test samples in the field for MIC will revolutionize oilfield microbiology and MIC management.
These solutions could provide a means to link molecular microbiological data to the physical and chemical environment in MIC management. Familiarity with case studies also can provide valuable information for interpreting molecular results. Books7 written by MIC experts can provide excellent resources studies and an overview of MMM approaches.
In 2007, the first International Symposium on Applied Microbiology and Molecular Biology of Oil Systems (ISMOS) assembled attendees from academia and industry to explore the application of emerging molecular methods to help solve challenges in oilfield systems such as MIC and reservoir souring. The most recent ISMOS meeting took place in San Diego, California, USA on June 6-9, 2017.
Coming up is CORROSION 2018 in Phoenix, Arizona, USA on April 15-19, where Technology Exchange Group (TEG) 187X will sponsor the technical symposium, “Microbiologically Influenced Corrosion,” and TEG 286X will sponsor the technical symposium, “Control of Problematic Microorganisms in Oil and Gas Field Operations.” Additionally, the 7th International Symposium on Applied Microbiology and Molecular Biology of Oil Systems (ISMOS-7) will take place on June 18-21, 2019 in Halifax, Nova Scotia, Canada. Conferences such as these provide an excellent opportunity to explore recent applications of emerging molecular tools through presentations, workshops, and poster sessions.
References
1 T.L. Skovhus, et al., “Practical Use of New Microbiology Tools in Oil Production,” SPE Offshore Europe Conference, paper no. 109104-MS (Richardson, TX: SPE, 2007).
2 T.L. Skovhus, R.B. Eckert, E. Rodrigues, “Management and Control of Microbiologically Influenced Corrosion (MIC) in the Oil and Gas Industry—Overview and a North Sea Case Study,” J. Biotechnol. 256 (2017): pp. 31-45.
3 NACE TM0212-2012, “Detection, Testing, and Evaluation of Microbiologically Influenced Corrosion on Internal Surfaces of Pipelines” (Houston, TX: NACE International, 2012).
4 T.L. Skovhus, R.B. Eckert, “Practical Aspects of MIC Detection, Monitoring and Management in the Oil and Gas Industry,” CORROSION 2014, paper no. 3920 (Houston, TX: NACE, 2014).
5 R.B. Eckert, T.L. Skovhus, “Advances in the Application of Molecular Microbiological Methods in the Oil and Gas Industry and Links to Microbiologically Influenced Corrosion,” International Biodeterioration & Biodegradation 126 (2018): pp. 169-176.
6 R.B. Eckert, T.L. Skovhus, “Using Molecular Microbiological Methods to Investigate MIC in the Oil and Gas Industry,” MP 50, 8 (2011).
7 T.L. Skovhus, D. Enning, J. Lee, Microbiologically Influenced Corrosion in the Upstream Oil and Gas Industry (Boca Raton, FL: CRC Press, 2017).
8 T.L. Skovhus, et al., “Introducing a Corrosion Management Approach to MIC Assessment, Control and Monitoring in the Oil and Gas Industry,” EUROCORR 2014, paper no. 7229 (Frankfurt am Main, Germany: DECHEMA, 2014).
9 J. Fichter, et al., “Use of a Methodological Panel to Identify the Source of Problematic Microbial Contamination in an Oil Shale Field,” CORROSION 2017, paper no. 9019 (Houston, TX: NACE, 2017).
This article was written by Amanda B. Chadee, Ph.D. from the University of Texas at Austin (Austin, Texas, USA), and Dr. Torben Lund Skovhus, researcher and project manager with VIA University College (Horsens, Denmark).